Diagnostic plots for hmclearn
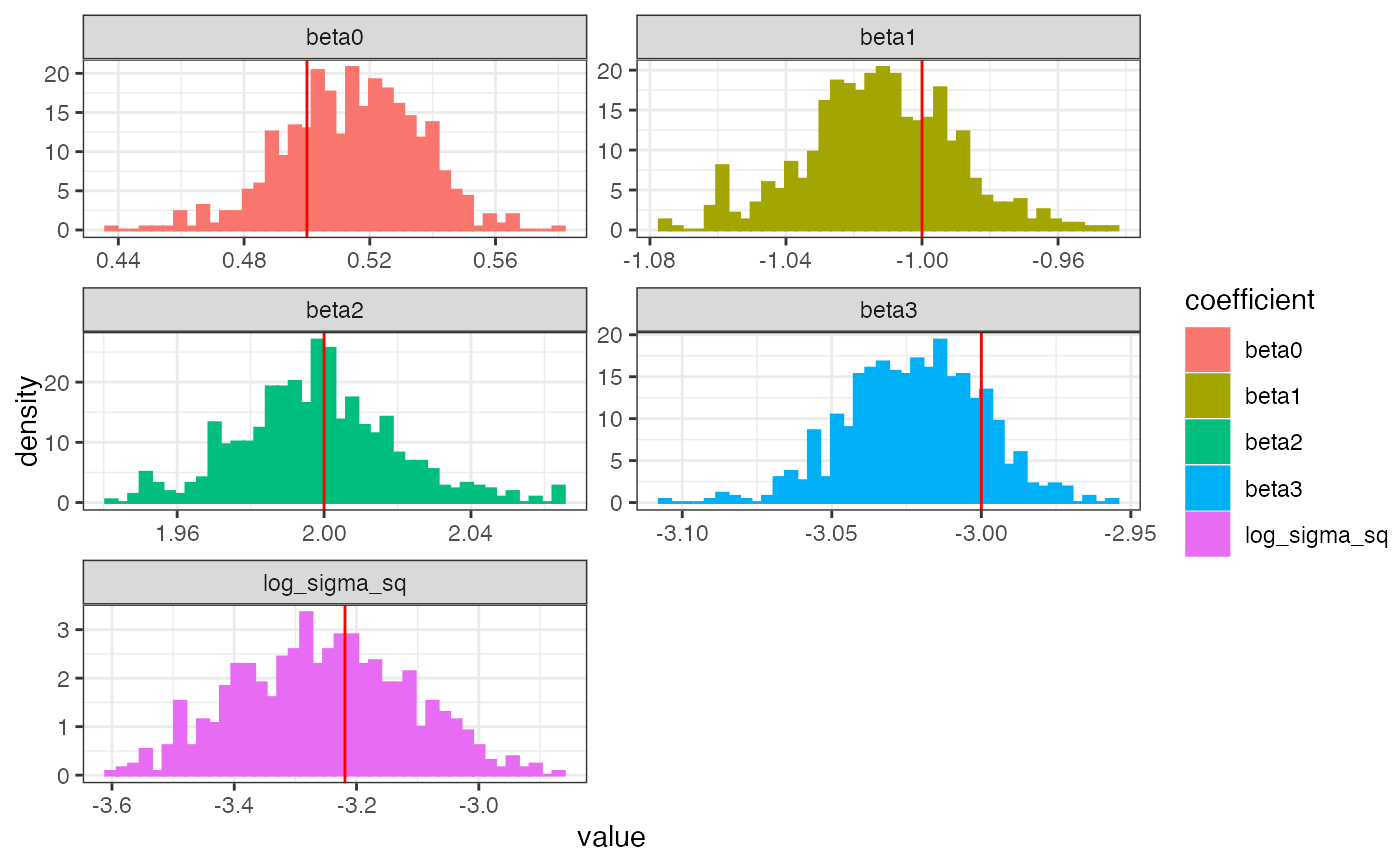
diagplots.hmclearn.RdPlots histograms of the posterior estimates. Optionally, displays the 'actual' values given a simulated dataset.
# S3 method for hmclearn diagplots( object, burnin = NULL, plotfun = 2, comparison.theta = NULL, cols = NULL, ... )
Arguments
| object | an object of class |
|---|---|
| burnin | optional numeric parameter for the number of initial MCMC samples to omit from the summary |
| plotfun | integer 1 or 2 indicating which plots to display. 1 shows trace plots. 2 shows a histogram |
| comparison.theta | optional numeric vector of parameter values to compare to the Bayesian estimates |
| cols | optional integer index indicating which parameters to display |
| ... | currently unused |
Value
Returns a customized ggplot object
Examples
# Linear regression example set.seed(522) X <- cbind(1, matrix(rnorm(300), ncol=3)) betavals <- c(0.5, -1, 2, -3) y <- X%*%betavals + rnorm(100, sd=.2) f <- hmc(N = 1000, theta.init = c(rep(0, 4), 1), epsilon = 0.01, L = 10, logPOSTERIOR = linear_posterior, glogPOSTERIOR = g_linear_posterior, varnames = c(paste0("beta", 0:3), "log_sigma_sq"), param=list(y=y, X=X), parallel=FALSE, chains=1) diagplots(f, burnin=300, comparison.theta=c(betavals, 2*log(.2)))#> $histogram#>